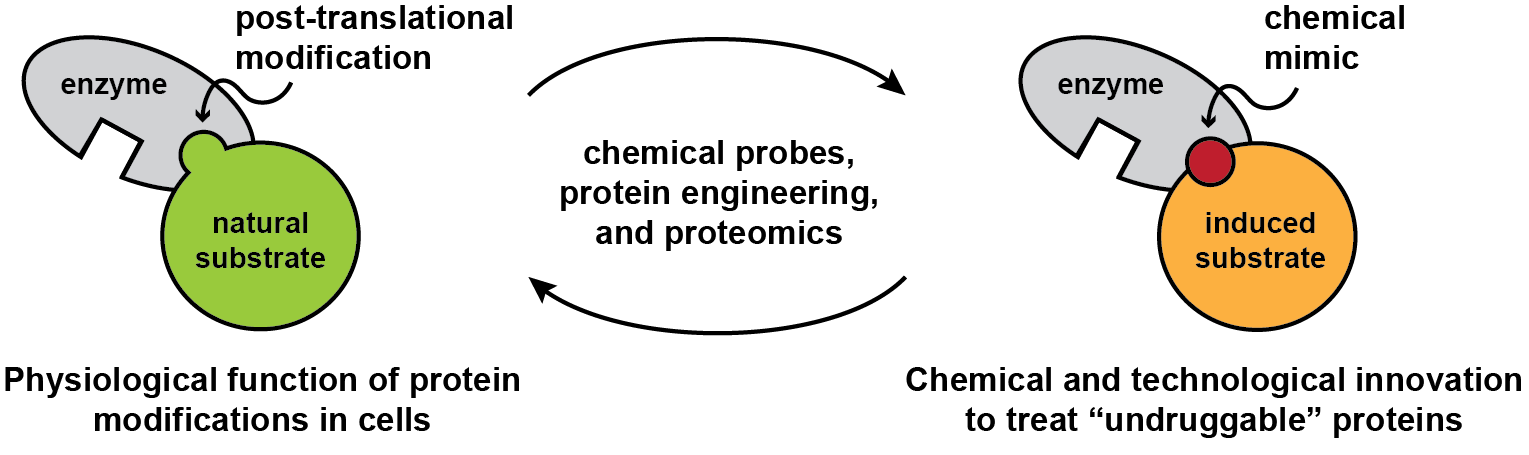
Reading and rewriting protein modifications
Our proteins are regulated by thousands of post-translational modifications that influence their structure and function. These modifications are mediated by enzymatic or non-enzymatic pathways and inhibit, enhance, or impart new functions to proteins. Research in the Woo lab aims to define and exploit the contribution of post-translational modifications to protein regulation. We develop chemical biology methods to elucidate protein modifications that are difficult if not impossible to study with traditional approaches and innovate strategies that allow us to precisely manipulate their functions, with important long-term implications for human health. In parallel, we study ligands that regulate protein function and seek to determine if these ligands are targeting natural regulatory sites in biological systems.
E3 ligase biology and protein damage response
The ubiquitin proteasome system uses E3 ubiquitin ligases to select protein substrates for ubiquitylation and protein degradation. Small molecule ligands can adapt ubiquitin ligases and chemically induce degradation of new target proteins. However, in many cases, the natural substrate recruitment mechanisms of E3 ligases are unknown, which makes the discovery of new chemical mimics challenging. We are studying these E3 ligases using chemoproteomic techniques and developing new ligands as probes for biological inquiry and strategies for therapeutic intervention. Recently, we used chemical approaches to discover the C-terminal cyclic imide as an overlooked form of protein damage that is recognized and removed by the E3 ligase adapter cereblon, an important target of the clinical therapeutics thalidomide and lenalidomide. This work has opened new directions in E3 ligase biology and enabled novel ligand discovery. We are now innovating approaches to investigate substrates of cereblon and identify readers for other overlooked chemical motifs in the proteome.
Key references:
Park S-M.*, Miyamoto D. K.*, Han G., Chan M., Curnutt N. M., Tran N. L., Velleca A., Kim J. H., Schurer A., Chang K., Xu W., Kharas M. G.#, Woo C. M.# “Dual IKZF2 and CK1α degrader targets acute myeloid leukemia cells.” Cancer Cell 2023, 41, 726.
Ichikawa S.*, Flaxman H. A.*, Xu W.*, Vallavoju N., Lloyd H. C., Wang B., Shen D., Pratt, M. R., Woo C. M. “The E3 ligase adapter cereblon targets the C-terminal cyclic imide degron.” Nature 2022, 610, 775.
Lin Z.*, Amako Y.*, Kabir F., Flaxman H. A., Budnik B., Woo C. M. “Development of photo-lenalidomide for cellular target identification.” J Am Chem Soc 2022, 144, 606.
Recorded Seminars
Northwestern University Proteostasis Consortium Seminar : “Investigation of protein damage modifications.” 26 February 2025. link to recording
Dana-Farber Cancer Institute Targeted Protein Degradation Webinar Series: “Chemical biology studies on the thalidomide-binding domain of cereblon.” 15 September 2022. link to recording
Writing and erasing O-GlcNAc from target proteins in cells
Modification of proteins with sugars produces glycoproteins, one of the essential chemical codes that cells use for nutrient sensing and signaling. O-linked N-acetyl glucosamine (O-GlcNAc) is found on Ser or Thr residues on thousands of nuclear and cytosolic proteins. However, functions of O-GlcNAc, how O-GlcNAc editors dynamically select these substrates, and why we need O-GlcNAc for life remain unanswered. To address these questions, we developed a nanobody-targeted O-GlcNAc writer and eraser for the systematic control of O-GlcNAc in live cells. We are applying our method to systematically investigate the impact of O-GlcNAc on protein function in live cells.
Key references:
Schwein P., Ge Y., Yang B., D’Souza A. K., Mody A., Shen D., Woo C. M. “Writing and erasing O-GlcNAc on casein kinase 2 alpha alters the phosphoproteome.” ACS Chem Biol 2022, 17, 1111.
Ramirez D. H., Yang B., D’Souza A. K., Shen D., Woo C. M. “Truncation of the TPR domain of OGT alters substrate and glycosite selection.” Anal Bioanal Chem 2021, 413, 7385.
Ge Y., Ramirez D. H., Yang B., D’Souza A. K., Aonbangkhen C., Wong S., Woo C. M. “Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase.” Nat Chem Biol 2021, 17, 593.
Ramirez D. H., Aonbangkhen C., Wu H. Y., Naftaly J. A., Tang S., O’Meara T. R., Woo C. M. "Engineering a proximity-directed O-GlcNAc transferase for selective protein O-GlcNAcylation in cells." ACS Chem Biol 2020, 15, 4, 1059.
Binding site mapping by chemical proteomics
We developed a chemical proteomics platform to elucidate the binding sites of small molecules to the global proteome, termed small molecule interactome mapping by photo-affinity labeling (SIM-PAL). SIM-PAL uses small molecule probes to capture and enrich binding sites in living systems, which we structurally elucidate by mass spectrometry. We have applied SIM-PAL to several bioactive small molecules and natural products to reveal their targets with high structural resolution on proteins in cells. Close investigation of the underlying photo-chemistry and development of new chemistries will further accelerate these target identification and mechanism of action studies.
Key references:
West A. V., Amako Y., Woo C. M. “Design and Evaluation of a Cyclobutane Diazirine Alkyne Tag for Photoaffinity Labeling in Cells.” J Am Chem Soc 2022, 144, 21174.
West A. V., Muncipinto G., Wu H. Y., Huang A. C., Labenski M. T., Jones L. H., Woo C. M. “Labeling preferences of diazirines with protein biomolecules.” J Am Chem Soc 2021, 143, 6691.
Flaxman H. A., Chang C. F., Wu H. Y., Nakamoto C. H., Woo C. M. “A Binding Site Hotspot Map of the FKBP12–Rapamycin–FRB Ternary Complex by Photoaffinity Labeling and Mass Spectrometry-Based Proteomics.” J Am Chem Soc 2019, 141, 11759.
Thank you to the following organizations for supporting our research:
Milton Fund, Merck Fellowship Fund, Dean’s Competitive Fund, Broad Institute.